Research Projects
The bacterial genus Streptococcus contains some of the most important pathogens and commensals of the human microbiome, with multiple species causing significant human morbidity and mortality. This project is revealing how three important Streptococcus pathogens utilize and modify host fatty acids and discover how this process integrates with quorum-sensing and virulence regulation. By defining the biochemical and signaling pathways connecting fatty acid utilization to pathogenic behavior, this work will uncover new molecular targets for prebiotic strategies that disarm pathogens.

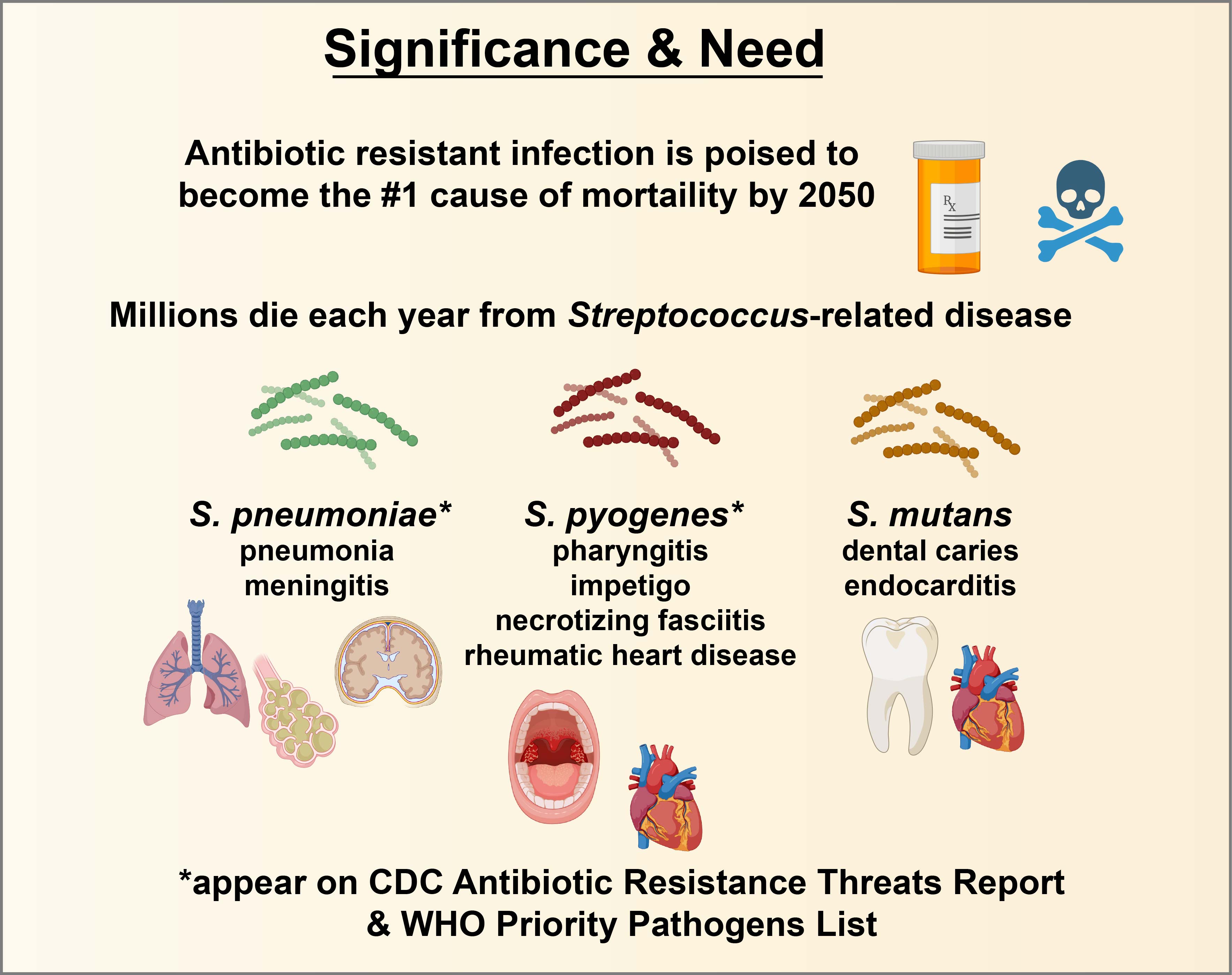
Resistance of pathogenic microbes to antibiotics is a growing worldwide concern, with global deaths due to antibiotic resistance predicted to overtake global deaths due to cancer and become the #1 cause of death worldwide by 2050. This project, funded by the Research Council of Norway, partners research labs from the Norwegian Institute of Public Health, the University of Oslo, TATA Consultancy Services (based in Delhi, India), the University of Campinas (Piracicaba, Brazil), McGill University (Montreal, Canada), the University of Illinois at Chicago, the Forsyth Institute, and OHSU to engage diverse trainees, clinicians, and scientists in educational outreach regarding the accelerating antibiotic resistance crisis and antibiotic stewardship.
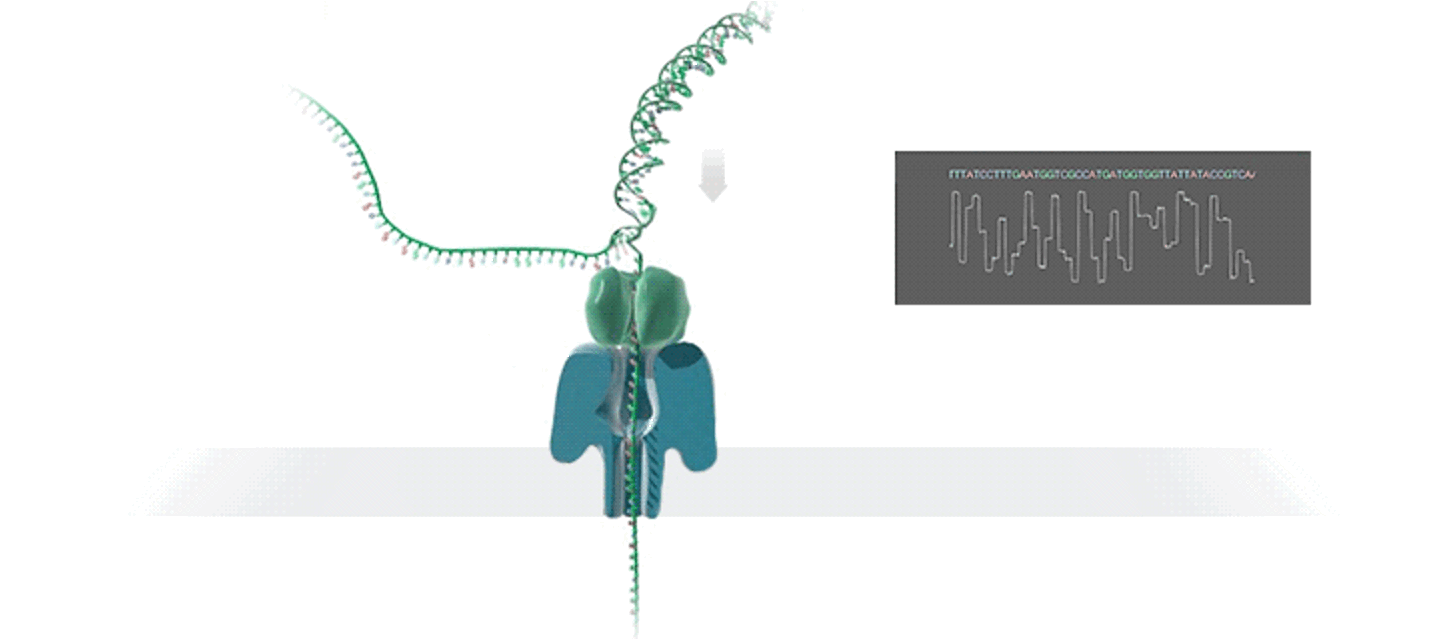
Nanopore sequencing technologies have gained considerable traction in recent years, with a critical inflection point in the sequencing industry being reached in 2022 when the Oxford Nanopore Technologies (ONT) sequencing platform acheived an accuracy on par with industry standard Illumina short-read sequencing. The Baker Lab was an early adapter of ONT technology, and in 2022 published the first protocols to obtain multiple complete bacterial genomes simultaneously directly from saliva using ONT sequencing. Are leveraging this technology to blaze new trails in oral microbiome research, specifically in regards to completing genomes of novel species, and examining the oral epigenome and epitranscriptome at scale.